DNA Replication
MCAT Biology - Chapter 1 - Section 2.2 - DNA - DNA Replication
- Home
- »
- MCAT Masterclass
- »
- Biological and Biochemical Foundations of Living Systems
- »
- Biology
- »
- DNA Replication – MCAT Biology
Sample MCAT Question - DNA Replication
Which of the following is true regarding the enzymes involved in DNA replication?
a) DNA helicase relieves strain in unwound DNA
b) Topoisomerase removes supercoils in DNA strands
c) DNA primase adds a short fragment of complementary DNA
d) DNA polymerase adds nucleotides in a 3’ to 5’ direction
B is correct. Topoisomerase removes supercoils in DNA strands.
The process of unwinding DNA during DNA replication creates strain. Topoisomerase removes supercoils in front of DNA helicase and relieves this strain. Answer choice A is incorrect. The enzyme helicase separates double-stranded DNA and forms two single-stranded DNA templates. Answer choice C is incorrect. DNA primase adds a short fragment of RNA complementary to the DNA template in order for DNA polymerase to begin to synthesize new DNA. Answer choice D is incorrect. DNA is always synthesized in the 5′ to 3′ direction.
Get 1-on-1 MCAT Tutoring From a Specialist
With MCAT tutoring from MedSchoolCoach, we are committed to help you prepare, excel, and optimize your ideal score on the MCAT exam.
For each student we work with, we learn about their learning style, content knowledge, and goals. We match them with the most suitable tutor and conduct online sessions that make them feel as if they are in the classroom. Each session is recorded, plus with access to whiteboard notes. We focus on high-yield topics if you’re pressed for time. If you have more time or high-score goals, we meticulously cover the entire MCAT syllabus.
DNA Replication Intro
DNA replication is the process of creating two identical DNA molecules from a single original DNA molecule. As discussed in a previous post, DNA replication is “semi-conservative”, meaning that the two resulting DNA molecules each contain a strand from the original DNA molecule. DNA replication is also an “all-or-none” process, meaning that once it starts, it will proceed to completion.
After reading this MCAT post, you should have a solid grasp of how DNA replication works and how it proceeds differently in prokaryotic and eukaryotic organisms.
Origins of Replication
The process of DNA replication begins at a site called the origin of replication, where an initiator protein will bind. It is important to note that the origin of replication between prokaryotes and eukaryotes differs. Prokaryotes have a circular chromosome that has one origin of replication (Figure 1). DNA replication starts at one position in the circular chromosome and works its way around the entire chromosome.

Eukaryotes, on the other hand, consist of multiple linear chromosomes, with multiple origins of replications on a single chromosome (Figure 2). Having so many origins of replication in eukaryotes helps to speed up the process of DNA replication.
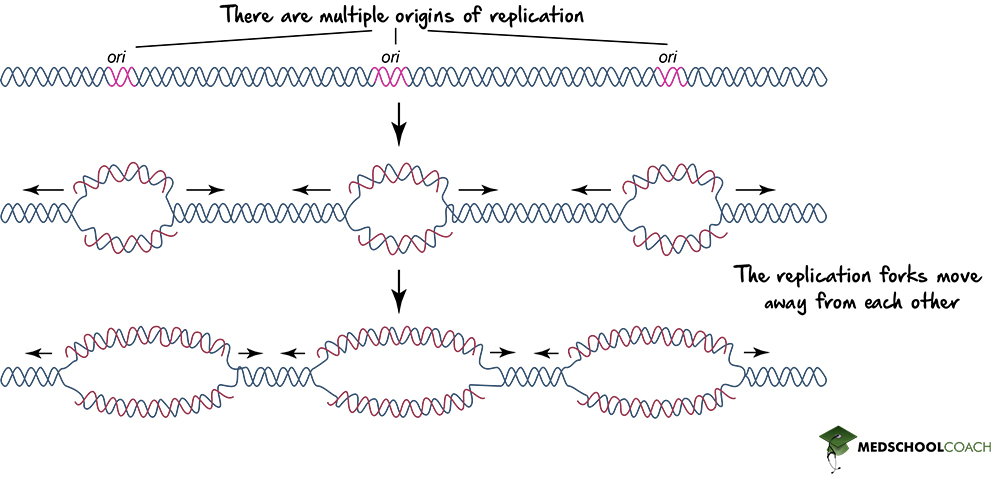
DNA Helicase, Topoisomerase, and the Replication Fork
As mentioned earlier, DNA replication first begins when an initiator protein binds to the origin of replication (Figure 3). When this happens, the initiator protein will unwind the DNA. However, apart from this basic function, it does not do much else. The first significant enzyme, however, is DNA helicase. DNA helicase binds to the double-stranded DNA and unwinds it to form a replication fork, creating two single-stranded DNA templates.
However, when DNA helicase separates the DNA into two strands, it creates strain in the DNA molecule. This strain or tension arises because of the double-helical structure of the DNA. When the DNA unwinds, sometimes it can overwind ahead of the replication fork and become tangled. Therefore, an enzyme called topoisomerase, or DNA gyrase, will cut through the phosphate backbone on one or both strands of DNA ahead of the replication fork. This action will help to untangle the DNA and relieve any built-up strain, allowing DNA helicase to continue unwinding the DNA. Afterward, the DNA backbone is once again reconnected.

DNA Primase and the RNA Primer
The next step involves an enzyme called DNA primase. Figure 4 illustrates how DNA primase binds to the leading strand of the DNA and lays down a short strand of RNA complementary to the single-stranded DNA template. This short fragment of RNA is called the RNA primer. The purpose of creating the RNA primer is so that DNA polymerase, which is the enzyme that synthesizes new fragments of DNA, can function. Because DNA polymerase needs an existing nucleotide with a 3′ OH group to which the next nucleotide can be added, DNA primase helps initiate the process of elongating the new nucleotide sequence. Also, it is important to note that DNA primase is making RNA and not DNA. In this way, the RNA primer will eventually have to be removed.
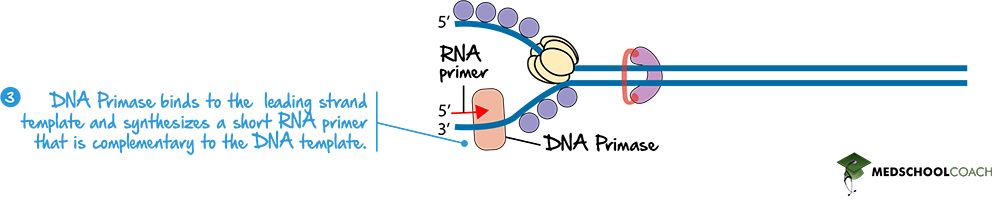
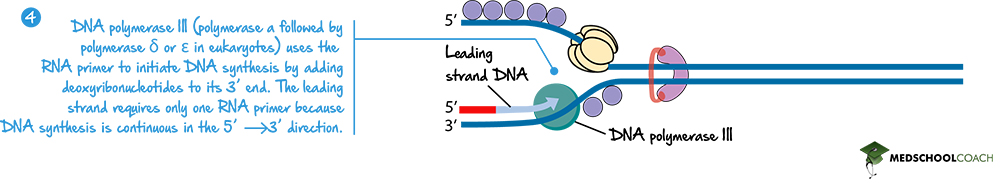
DNA Polymerase and the Leading Strand
Once the RNA primer is present, DNA polymerase can function. There are several types of DNA polymerase enzymes in both prokaryotes and eukaryotes, and it is not important to understand each of their specific functions for the MCAT exam. Figure 5 shows DNA polymerase III, which is the primary DNA polymerase in prokaryotes.
DNA polymerase works by using the template strand of DNA and adding DNA nucleotides in the 5’ to 3’ direction. The strand it is creating is called the leading strand. Essentially, as DNA helicase unwinds the DNA molecule, the leading strand is being continuously synthesized. Nevertheless, since DNA is antiparallel, and DNA polymerase adds nucleotides in the 5’ to 3’ direction, the opposite strand travels in the 3’ to 5’ direction. DNA polymerase can only work in the 5’ to 3’ direction, so the opposite strand, known as the lagging strand, is synthesized differently.
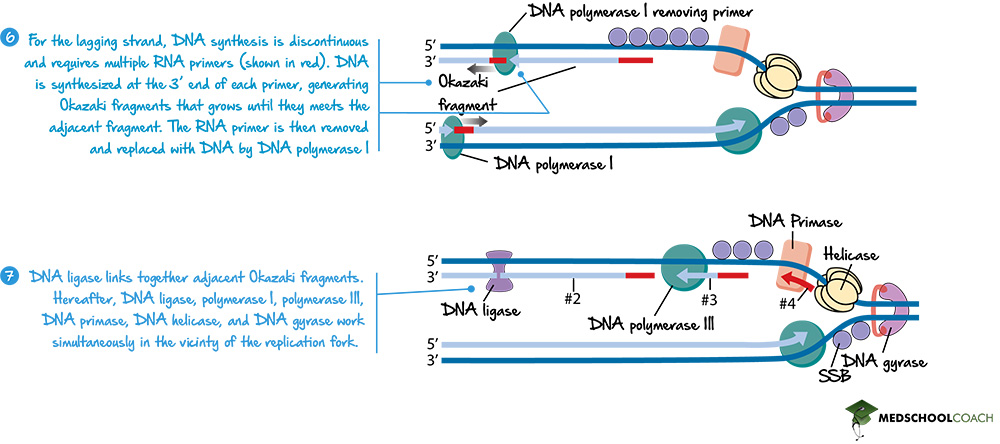
The Lagging Strand and Okazaki Fragments
In terms of the lagging strand, it involves multiple RNA primers. As DNA helicase unwinds the DNA, DNA primase creates an RNA primer near the replication fork. DNA polymerase III can create a new strand of DNA using that primer in 5’ to 3’ direction (Figure 6). However, as helicase continues to unwind the strand of DNA, DNA primase has to create a new RNA primer once again. DNA polymerase III once again creates a new strand of DNA in the 5’ to 3’ direction until it reaches the fragment of DNA it created before. In this way, the lagging strand is created in fragments, known as Okazaki fragments.
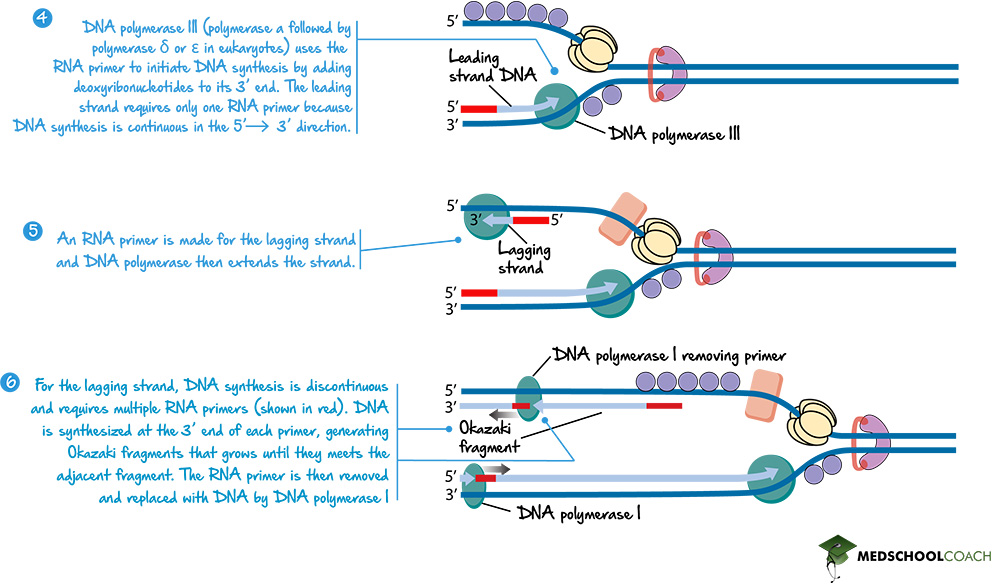
DNA Polymerase I and DNA Ligase
Lastly, the RNA primer, as previously mentioned, has to be removed. In prokaryotes, this is done by DNA polymerase I, which will replace the RNA primer with DNA. Figure 7 shows how DNA polymerase I removes the primer. However, it is essential to note that when RNA is replaced with DNA, the gap between the Okazaki fragments will be left behind. Another enzyme, DNA ligase, will ligate or seal the gaps between the Okazaki fragments (Figure 7).
DNA Polymerase Proofreading Activity
An important note about the enzyme DNA polymerase is that it has proofreading activity. Proofreading activity is a way that DNA polymerase can correct its mistakes. For example, if the wrong base pair is added within the genetic sequence, this could lead to mutations in the offspring and severe adverse effects. Generally, then, organisms try to prevent mutations when possible. In this way, as DNA polymerase is synthesizing new fragments of DNA, it proofreads what it is adding. If it adds something incorrectly, it can replace it right away and correct its error. Therefore, errors in DNA replication happen at a very low frequency.
Explore More MCAT Masterclass Chapters
Take a closer look at our entire MCAT Masterclass or explore our Biochemistry lessons below.

One-on-One Tutoring
Are you ready to take your MCAT performance to a whole new level? Work with our 99th-percentile MCAT tutors to boost your score by 12 points or more!
See if MCAT Tutoring can help me
Talk to our enrollment team about MCAT Tutoring

MCAT Go Audio Course
Engaging audio learning to take your MCAT learning on the go, any time, any where. You'll be on the way to a higher MCAT score no matter where you are. Listen to over 200+ lessons.
MCAT Practice Exams
Practice makes perfect! Our mock exams coupled with thorough explanations and in-depth analytics help students understand exactly where they stand.
MCAT Prep App
Access hundreds of MCAT videos to help you study and raise your exam score. Augment your learning with expert-created flashcards and a question banks.